Recently, scientists published a research paper titled “UDA-seq: universal droplet microfluidics-based combinatorial indexing for massive-scale multimodal single-cell sequencing” in Nature Methods.

The research team independently developed a novel single-cell multi-omics sequencing technology called UDA-seq. By optimizing and upgrading the widely used droplet microfluidics platform with a combinatorial indexing approach, they achieved the “decoupling” of cell throughput and false singlet rates, overcoming throughput limitations.
UDA-seq is a universal strategy for enhancing throughput, supporting common single-cell multi-omics applications such as RNA and VDJ co-detection, RNA and ATAC co-detection, and RNA and CRISPR perturbation co-detection within the same cell. This study achieved a record-breaking single-channel cell throughput of over 100,000 cells, a 10- to 20-fold improvement over existing technologies, while maintaining compatibility with multimodal analyses. Additionally, the researchers employed genetic diversity combined with bioinformatics to enable label-free single-channel multiplexing of 20 to 40 human samples, significantly reducing per-sample data costs. The study also established a solution for obtaining high-quality, multimodal, single-cell precision data from small frozen blood and biopsy samples on a large scale.
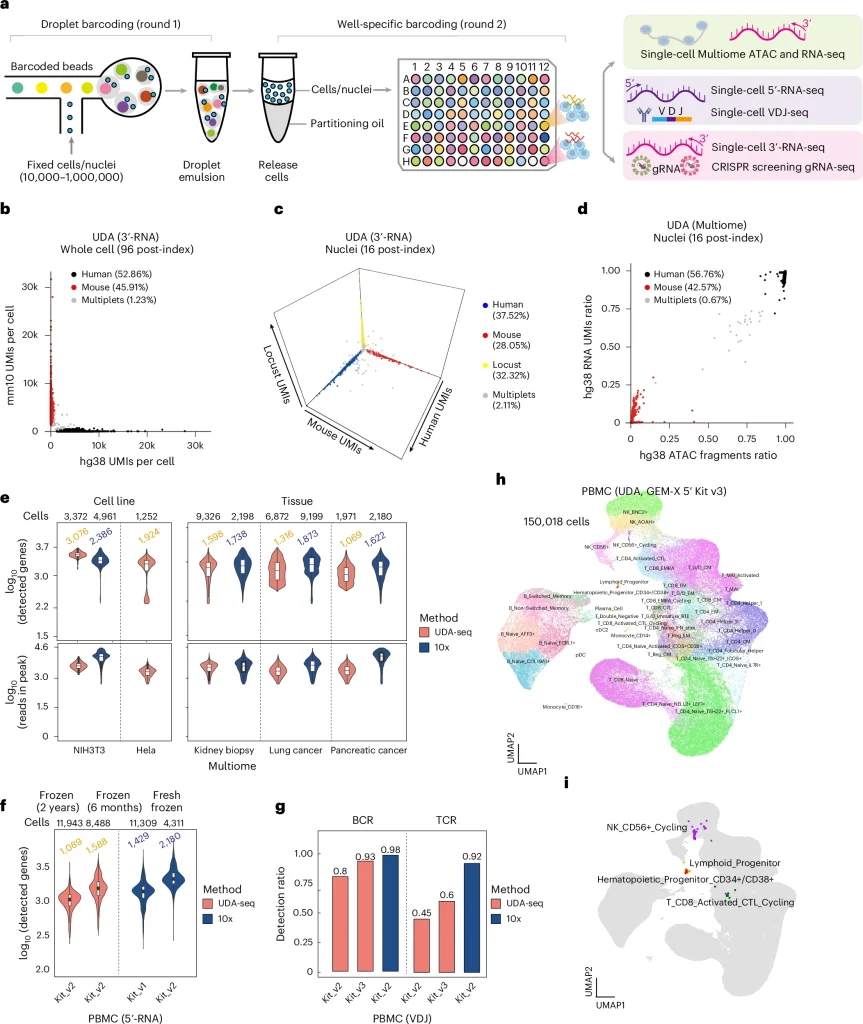
The workflow of UDA-seq and its performance across different modalities and samples
Furthermore, the study applied UDA-seq in three scenarios: aging, disease population cohort studies, and high-throughput CRISPR screening research. For kidney tissues from healthy individuals and patients with various complex kidney diseases, the study constructed a single-cell RNA and ATAC dual-omics atlas. The research overcame the challenges of processing small frozen samples, requiring only one-fifth of a needle-biopsy sample per case. In droplet microfluidics, the team collected 200,000 high-quality dual-modal single-cell data from 35 donors using two-channel reactions. By developing new algorithms, the study identified rare cell types, including podocytes (POD) and endothelial-glomerular cells (EC-GC), associated with proteinuria and kidney injury phenotypes, as well as the regulatory networks and key factors of critical cell subpopulations.
Using UDA-seq, the study constructed a PBMC single-cell transcriptomic and immune receptor sequencing dual-omics atlas for a naturally aging population cohort. With two-channel reactions, 120,000 high-quality single-cell data were collected, revealing aging-related cell subtypes and identifying a new aging-related cell subtype—ITGB1+PREX1+ Naive CD4+ T cells. UDA-seq also demonstrated its compatibility with high-throughput CRISPR screening, analyzing the cellular responses to perturbations of the bromodomain protein gene family in gastric cancer cell lines.
This study breaks through existing technical bottlenecks, enhancing single-cell sequencing throughput while maintaining compatibility with multimodal analyses. It demonstrates reliability and high quality in real-world samples, achieving low-cost and efficient data production. The work developed new algorithms to identify key cell subpopulations, regulatory networks, and critical factors associated with clinical phenotypes, establishing a novel research paradigm for large-scale studies of health and disease cohorts. Additionally, the study upgraded the commonly used droplet microfluidics platform, which holds potential to support data generation for next-generation multimodal virtual cell-based large models.
Reference:
Li, Yun et al. “UDA-seq: universal droplet microfluidics-based combinatorial indexing for massive-scale multimodal single-cell sequencing.” Nature Methods, 10.1038/s41592-024-02586-y. 20 Jan. 2025, doi:10.1038/s41592-024-02586-y
Related Services:
Microfluidic Development Services for Droplet Generator and Flow Chemistry