This article provides a comprehensive overview of the progress made in the application of droplet microfluidics to single-cell analysis. The development of droplet microfluidics and its key features as a single-cell tool are described, while the complexity of cellular regulation and gene expression patterns at the individual cell level is also expressed based on existing studies In addition, the emerging field of cell-to-cell and cell-microbial interactions is explored, revealing the complexity of inter-cellular communication and symbiotic relationships, and providing insights into a deeper understanding of cellular heterogeneity and function.
What is Droplet Microfluidics?
Microfluidic droplet technology uses two or more immiscible fluids as the dispersed phase and the continuous phase, respectively. The dispersed phase is split by the continuous phase into tiny droplets, which are formed spontaneously at the interface of the two phases under the combined effect of surface tension and shear.
In droplet microfluidic systems, three main channel structures are commonly used to generate droplets: T-crossing structures, flow-focusing structures, and coaxial co-flow structures. The droplets are mainly categorized into W/O (water-in-oil) and O/W (oil-in-water). In addition, the multiple emulsions mentioned are complex polydisperse systems where W/O and O/W coexist.
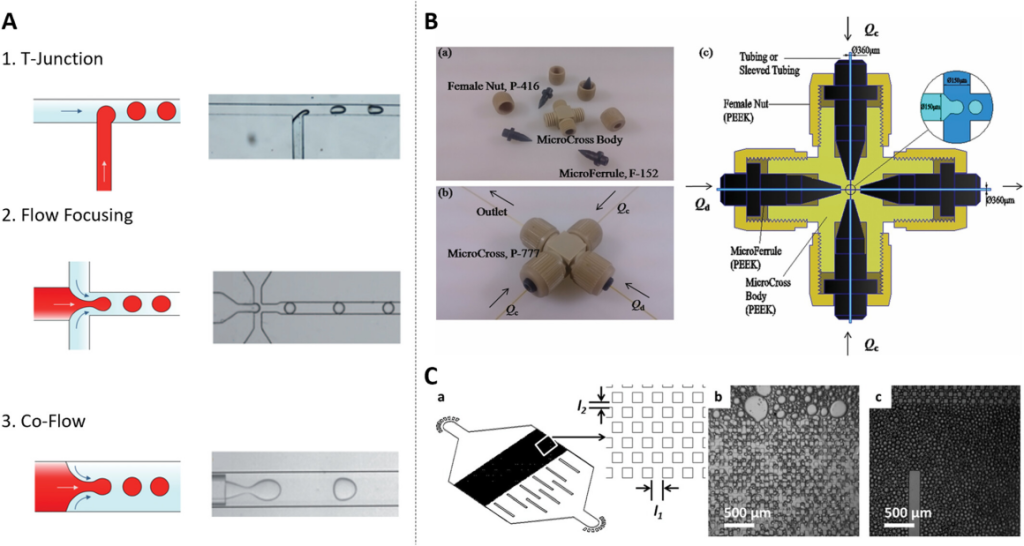
Fig. 1 Schematic diagram of droplet microfluidic structure.1,2
With further research, Wu et al. introduced a new 3D Micro-Cross droplet generation structure that is easy to assemble. Compared with traditional methods, firstly, the Micro-Cross can be seamlessly integrated with the droplet detection device, and the generated droplets can be collected and processed directly inside the tube. Secondly, the inner wall can generate W/O and O/W droplets without any wetting treatment. Furthermore, the three-dimensional design of the Micro-Cross allows for droplet separation in any direction, making it even more versatile. In addition, the droplet generation frequency of this method exceeds that of conventional PDMS (two-dimensional polydimethylsiloxane) chips, which is another major advancement in the field of droplet microfluidics.
Cell Encapsulation in Droplet Microfluidics
Over the past decade, scientists have been investigating the creation of various microfluidic materials and devices to improve the encapsulation of living cells. Viovy et al. proposed a simple hydrodynamic method in which droplets encapsulating single cells spontaneously self-sort from smaller empty droplets as the cells enter a narrow water jet created by a flow-focusing device to encapsulate themselves.
Toner et al. proposed a new method to self-organize cells into two uniformly spaced flow bundles before the emulsification process.
In 2019, Weitz et al. designed surfactants that have four hydroxyl groups in their polar heads, which enhance hydrogen bonding and provide better high-temperature stability and small-molecule retention compared to PEG-PFPE surfactants. It plays an integral role in experiments requiring droplet stabilization integrity and compatibility with encapsulated biomolecules.
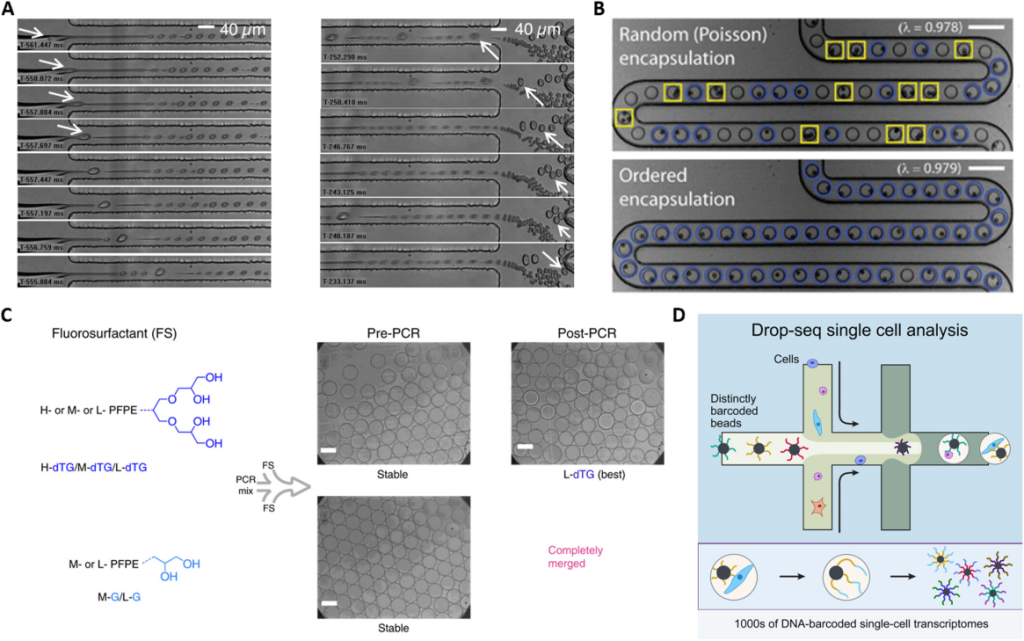
Fig. 2 Microfluidic droplet-based labeling for single-cell encapsulation, droplet stability, and single-cell analysis.1,2
In droplet microfluidics each droplet is a unique reactor, and when millions of single-cell droplets are generated, how to identify each distinct single cell? Weitz et al. reported a platform that combines droplet microfluidics with DNA oligonucleotide sequences, also known as DNA barcodes, to enable genome-wide expression profiling at single-cell resolution. This method, called Drop-seq, encapsulates thousands of single cells into nanoliter-sized droplets, and by combining a unique barcode with each cell’s RNA, Drop-seq can analyze thousands of single-cell mRNA transcripts simultaneously while retaining information about each transcript. Drop-seq represents a significant advance in biological research, facilitating single-cell resolution for routine transcriptional profiling.
Droplet Microfluidics for Single Cell Analysis
The advent of droplet microfluidics has revolutionized single-cell transcript sequencing and driven the development of new platforms for single-cell DNA and RNA sequencing. Since the Weitz team introduced droplet-based, highly parallel single-cell transcript sequencing in 2015, the technology has become the gold standard in the field. It has prompted a surge in single-cell data mining in recent years, providing unprecedented insights across a wide range of biological disciplines, including developmental biology, cancer biology, and microbiology. The ability to distinguish and quantify transcripts from thousands of single cells in a population has transformed our understanding of complex tissues and cellular functions.
The traditional steps for single-cell genetic analysis include single-cell droplet encapsulation, lysis of the droplet to extract DNA, and subsequent amplification experiments and detection. In contrast, Kumaresan et al. pioneered the use of a droplet microfluidic device to encapsulate single cells in agarose solution, which gels after cooling, and then separates them from the oil phase and lyses the cells using a mixture of sodium dodecyl sulfate (SDS) and proteinase K. PCR amplification is performed to label the target genetic material in the droplet for subsequent detection.
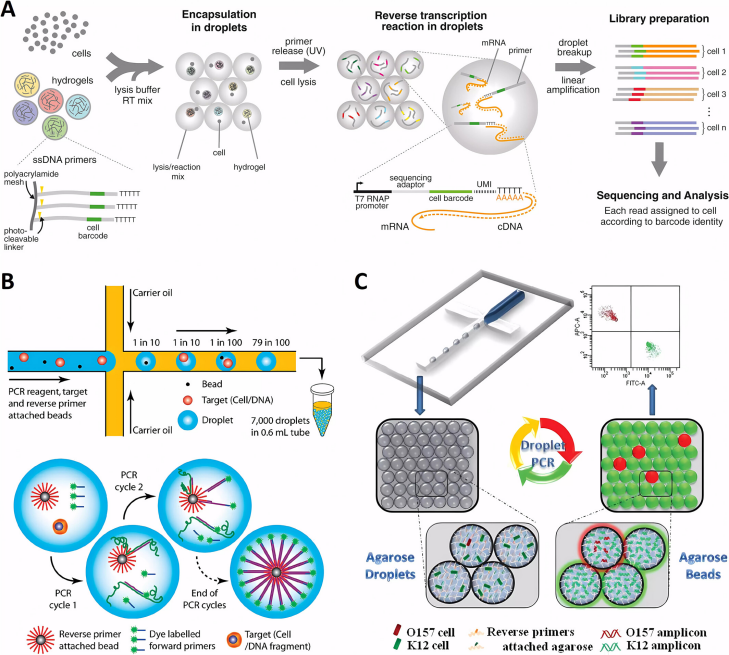
Fig. 3 Microfluidic systems for PLGA drug delivery systems.1,2
The precise pairing of single cells with magnetic beads in the droplet is an extremely important step and a technical difficulty. To solve this problem, Yang et al. developed a microfluidic technique for emulsion PCR using agarose droplets without magnetic beads, with reverse transcription primers covalently attached directly to the agarose. This approach increased droplet production efficiency by one to two orders of magnitude. The agarose remained liquid at all PCR cycling temperatures and its efficiency exceeded that of 95% of PCR reactions. Using this technique, single-cell PCR was successfully performed in the context of over 100,000 normal K12 cells to identify a single pathogen, Escherichia coli O15:H7.
The method was also optimized for use in single-cell RT-PCR experiments by injecting cells and RT-PCR reagents from a separate aqueous inlet. Single-cell RT-PCR analysis revealed significant differences in the expression levels of the cancer biomarker gene EpCAM in different cancer cell types at the single-cell level. This new method is expected to be applied to viral RNA detection and further single-cell transcription studies, paving the way for breakthroughs in understanding gene expression heterogeneity.
Creative Biolabs’ Microfluidic Platform
The article systematically summarizes significant advances in droplet microfluidics for single-cell analysis. These breakthroughs have made it possible to obtain high-resolution data from single cells, offering great promise for exploring cellular activity. Although substantial progress has been made in the last decade, there is still great potential for further development, especially in terms of throughput and generalizability. In addition, the introduction of artificial intelligence and machine learning techniques has provided new perspectives on data processing and analysis, taking single-cell research to a new level.
Creative Biolabs’ microfluidics service platform leverages advanced lab-on-a-chip technologies to streamline fluid manipulation at the microscale, delivering unmatched precision, throughput, and integration. Whether you’re exploring single-cell analysis, droplet microfluidics, or organ-on-a-chip models, our team of experts can guide your project from conceptual design to functional prototype—and beyond.
References
- Liu, Chang, and Xiaoyu Xu. “Droplet Microfluidics for Advanced Single‐Cell Analysis.” Smart Medicine2 (2025): e70002. https://doi.org/10.1002/smmd.70002
- Distributed under Open Access license CC BY 4.0, without modification.