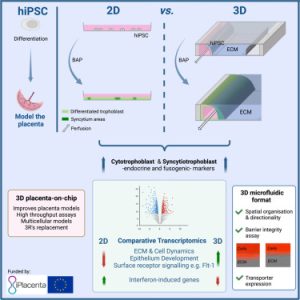
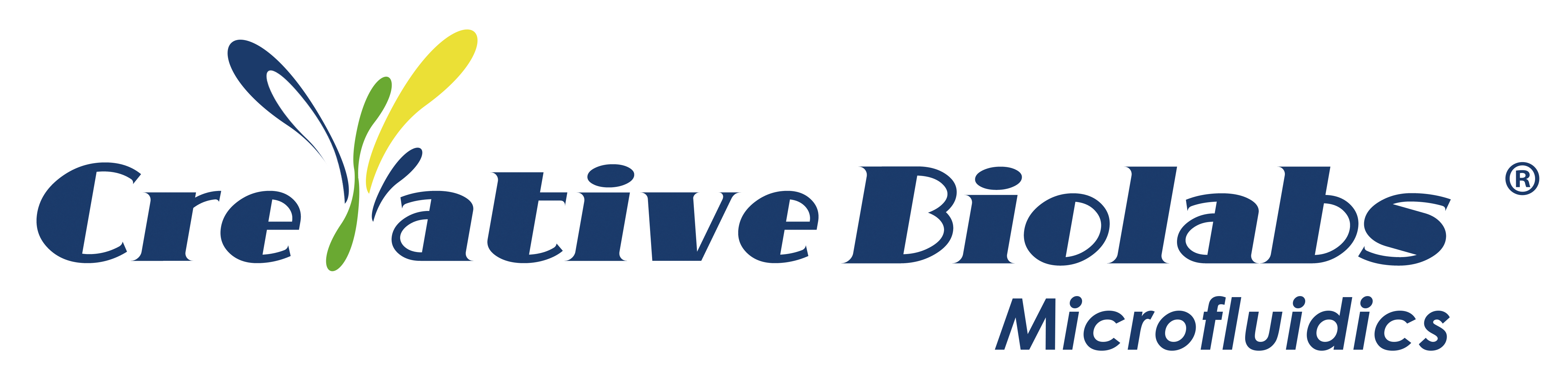On July 13, 2023, a research team from the University of Dundee School of Medicine in the UK published a research paper titled “Development of a Human iPSC-Derived Placental Barrier-on-Chip Model” in the journal iScience. This study utilizes hiPSC-derived trophoblast cells to develop a physiologically relevant and scalable placental barrier chip model, named iPlacenta. The researchers cultured induced pluripotent stem cell-derived trophoblast cells in the device. Following differentiation, trophoblast cells self-assembled into intricate 3D structures, forming robust structural barriers, and expressed genes associated with early placental development. This study demonstrated the feasibility of generating differentiated primitive syncytia using hiPSCs on a microfluidic platform, promising significant advancements in placental barrier models and their applications in pregnancy research and therapeutic evaluation.

Validation of Trophoblast Differentiation Protocol in 2D Culture Conditions
In this study, the researchers utilized ChiPS4, a hiPSC cell line derived from primary human dermal fibroblasts, and unconditioned mTeSR1 medium for cell maintenance and BAP differentiation—a unique combination not reported elsewhere for trophoblast differentiation. The trophoblast differentiation protocol was meticulously validated by assessing morphological characteristics and measuring molecular markers over 6 days of BAP exposure. This involved a comprehensive analysis of pan-trophoblast, CTB, EVT, and STB-specific markers (refer to Fig. 1). Phase contrast images of BAP-treated hiPSCs revealed a rapid loss of pluripotent cell phenotype within 48 hours, transitioning into a flattened epithelial-like morphology (see Figure 1B). The BAP-exposed cells formed a confluent monolayer within 48 hours, later becoming hyperconfluent and forming cell aggregates detached from the cell layer, consistent with previous reports on hESC-derived trophoblast cells (see Figure 1B). These results confirmed that the modified BAP protocol effectively committed ChiPS4 cells to the trophoblast lineage, generating a mixed population of CTB, STB, and EVT over 6 days of differentiation.
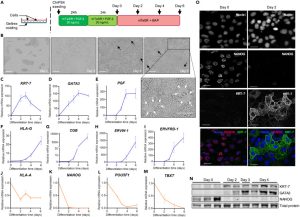 Figure 1. Validation of Trophoblast Differentiation Protocol in 2D Culture Conditions
Figure 1. Validation of Trophoblast Differentiation Protocol in 2D Culture Conditions
Generation of hiPSC-Derived Trophoblast Cells in 3D Microfluidic Devices
The researchers differentiated ChiPS4 cells directly within the OrganoPlate 3-channel microfluidic chip using a BAP differentiation protocol similar to 2D culture conditions. After 4 days, the differentiated ChiPS4 cells spontaneously assembled into a hollow tubular structure (see Figure 2B, 2C). These tubular structures expressed positive trophoblast markers KRT-7, GATA3, and PGF while downregulating pluripotency and somatic markers, confirming the generation of trophoblast cells. Notably, these cells exhibited invasive behavior, entering the collagen gel layer. Further RNA sequencing analysis validated the differentiation toward trophoblast cells. Compared with traditional two-dimensional culture conditions, the microfluidic environment provided a more standardized cell layout, forming a tight barrier within the tubular structure. In summary, the microfluidic environment facilitated the directed differentiation of ChiPS4 into functional trophoblast cells, making it an ideal system for establishing a trophoblast model closely resembling in vivo conditions.
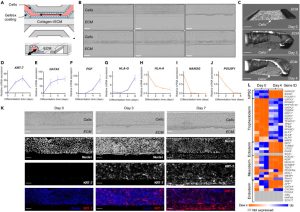 Figure 2. Generation of hiPSC-Derived Trophoblast Cells in a 3D Microfluidic Device
Figure 2. Generation of hiPSC-Derived Trophoblast Cells in a 3D Microfluidic Device
On-Chip Trophoblast Differentiation of hiPSCs Generates Functional Syncytia
The data analysis from the single-cell profiling panel revealed the presence of mature syncytiotrophoblasts (STB) in cells treated with BAP for 4 days. The expression of STB markers, such as hCG, enzymes involved in estrogen and progesterone biosynthesis, and pregnancy-specific glycoprotein (PSG) were significantly upregulated. Additionally, the pro-fusion genes syncytin-1 and syncytin-2 were also upregulated, indicating the presence of thin cytotrophoblasts (CTB) fused into STB (see Fig. 3). RT-qPCR results demonstrated that the expression timeline of ERVW-1, ERVFRD-1, and CGB was consistent with the immunofluorescence results. Immunofluorescence analysis showed that β-hCG was primarily expressed on the collagen side, and E-cadherin expression decreased in this area, confirming the fusion of cells into a syncytium. A 3D reconstruction of confocal images confirmed progressive β-hCG induction targeting the extracellular matrix (ECM) (see Figure 3J). This model effectively replicates key physiological functions of human syncytiotrophoblasts and can be instrumental in studying early implantation and trophoblast development. Collectively, these data demonstrate that hiPSC-derived mixed trophoblast populations, obtained in a microfluidic platform, contain fully differentiated functional STBs with fusion and endocrine activity.
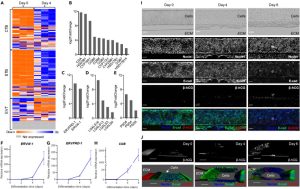 Figure 3. On-Chip Trophoblast Differentiation of hiPSCs Produces Functional Syncytia
Figure 3. On-Chip Trophoblast Differentiation of hiPSCs Produces Functional Syncytia
Evaluation of a hiPSC-Derived Placenta-on-a-Chip Model for Placental Barrier Transport Studies
The structural integrity of hiPSC-derived trophoblast tubular structures in the microfluidic platform was evaluated assessing their permeability to compounds of various molecular sizes. By day 2, both fluorescent compounds were leaking from the cell container in all measured chips (see Figures 4A-4D). From day 4 to day 6, there was a complete lack of fluorescence signal 10 minutes after the introduction of dextran in the gel zone, indicating the formation of a tight leakage barrier against 155 kDa and 10 kDa compounds. RNA-seq and differential gene expression analysis revealed significant upregulation of 10 ABC transporters and 66 SLC transporters. Notable upregulated drug transporters included the placental organic anion transporter OAT-4 and the organic cation/carnitine transporter OCTN2 (refer to Fig. 4E). This model exhibits promising potential for placental transport studies.
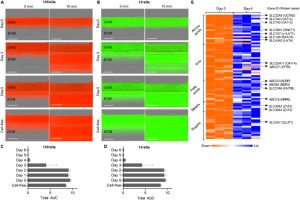 Figure 4. Evaluation of hiPSC-Derived Placenta-on-a-Chip Models for Placental Barrier Transport Studies
Figure 4. Evaluation of hiPSC-Derived Placenta-on-a-Chip Models for Placental Barrier Transport Studies
Microfluidic Culture Alters the Transcriptomic Profile of hiPSC-Derived Trophoblast Cells
Comparison of genome-wide expression profiles of hiPSC-derived trophoblast cells differentiated on microfluidic devices and conventional 2D plates for 4 days revealed interesting findings (seeFigure 5B). Among the 1019 differentially expressed genes, most upregulated genes were related to the reorganization of the extracellular matrix (ECM), such as collagen assembly and ECM organization. Additionally, some biological pathways associated with surface receptors and signal transduction were influenced by microfluidic culture, activating the receptor tyrosine kinase signaling pathway and inhibiting the type I interferon signaling pathway. Functional enrichment analysis indicated that up-regulated genes were related to processes such as ECM organization, epithelial development, and drug response, while down-regulated genes were related to stress and inflammatory responses. These results suggest that the microfluidic environment promotes syncytialization and differentiation of trophoblast cells by activating cell-ECM interactions, mechanosensing, and surface receptor signaling.
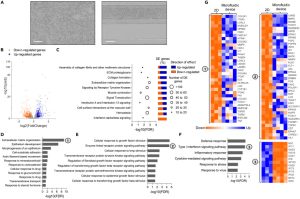 Figure 5. Customized Microfluidic Culture Alters the Transcriptome Profile of hiPSC-Derived Trophoblast Cells
Figure 5. Customized Microfluidic Culture Alters the Transcriptome Profile of hiPSC-Derived Trophoblast Cells
Summary and Discussion
This study aimed to differentiate human induced pluripotent stem cells into trophoblast cells and culture them within a porous microfluidic chip device, developing a more physiologically relevant and scalable placental barrier model. The researchers first validated that BAP treatment can effectively differentiate ChiPS4 hiPSCs into a mixed population of accumulating trophoblasts, syncytiotrophoblasts, and extracellular trophoblasts under two-dimensional culture conditions. Subsequently, the same BAP differentiation protocol was implemented directly within a microfluidic chip, leading hiPSC-derived trophoblast cells to spontaneously assemble into a three-dimensional tubular structure. These cells exhibited invasive behavior and expressed markers of functional syncytia. This tubular trophoblast structure formed a barrier impermeable to fluorescent tracers, indicating structural integrity. Furthermore, it expressed placenta-specific transporters such as GLUT1 and BCRP, suggesting its potential for transport studies. Transcriptomic comparison highlighted that microfluidic culture enhanced cell-matrix interactions, mechanosensitivity, and syncytialization processes. In conclusion, this study demonstrates the feasibility of differentiating hiPSCs into functional trophoblast cells directly within a microfluidic device, creating improved placenta-on-a-chip models for scientific research and drug screening applications. Combining hiPSC-derived trophoblast cells with a perfused porous format compensates for the limitations of existing placental barrier models.
Reference:
Lermant A, Rabussier G, Lanz HL, Davidson L, Porter IM, Murdoch CE. Development of a human iPSC-derived placental barrier-on-chip model. iScience. 2023 Jul 13;26(7):107240. doi: 10.1016/j.isci.2023.107240. PMID: 37534160; PMCID: PMC10392097.
Related Services:
Cell Culture and Organ-On-A-Chip Model Development Services
CBLMF™ 3D Tissue and Organ-On-A-Chip Model Development Services